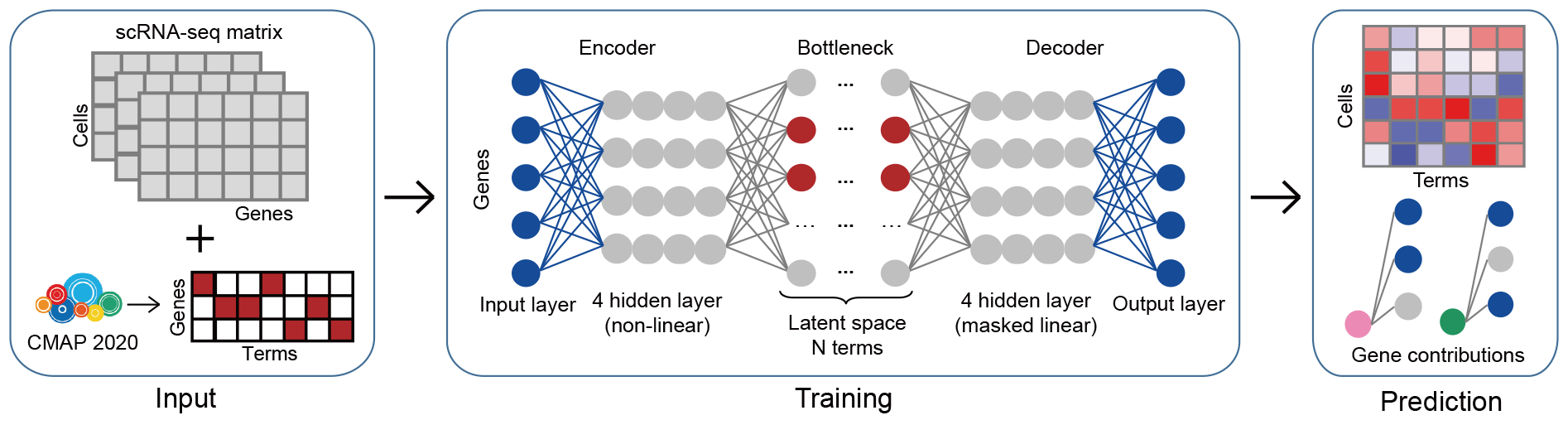
A deep learning framework for in silico screening of anticancer drugs at the single-cell level.
We introduce a deep learning framework named Shennong for in silico screening of anticancer drugs for targeting each of the landscape cell clusters. Utilizing Shennong, we could predict individual cell responses to pharmacologic compounds, evaluate drug candidates’ tissue damaging effects, and investigate their corresponding action mechanisms. Prioritized compounds in Shennong’s prediction results include FDA-approved drugs currently undergoing clinical trials for new indications, as well as drug candidates reporting anti-tumor activity. Furthermore, the tissue damaging effect prediction aligns with documented injuries and terminated discovery events. This robust and explainable framework has the potential to accelerate the drug discovery process and enhance the accuracy and efficiency of drug screening.
The training and prediction results could be obtained and queried on our website (http://bis.zju.edu.cn/shennong/index.html). Processed count matrices and cell annotations are available at figshare (https://doi.org/10.6084/m9.figshare.25497445).
Citation: Peijing Zhang†, Xueyi Wang†, Xufeng Cen†, Qi Zhang†, Yuting Fu, Yuqing Mei, Xinru Wang, Renying Wang, Jingjing Wang, Hongwei Ouyang, Tingbo Liang*, Hongguang Xia*, Xiaoping Han*, and Guoji Guo*. A deep learning framework for in silico screening of anticancer drugs at the single-cell level. National Science Review, 2025, 12(2):nwae451. DOI: https://doi.org/10.1093/nsr/nwae451.
Python packages
scanpy >= 1.9.2
scarches >= 0.5.7
torch >= 1.13.1
pandas >= 1.5.3
numpy >= 1.23.5
gdown >= 4.6.3
The scripts command_example.sh or example.ipynb shows how to predict individual cell responses to pharmacologic compounds with Shennong framework. The visualization of training and prediction results can be found in example.ipynb.
- scRNA data
- Perturbation data
perturbation.gmt perturbation.gmt.h5ad (high-confidence signatures of CMap with already preproceessed)
python ../script/preprocess.py adata_train.h5ad ../data/perturbation_test.gmt ../data/perturbation_test.gmt.h5ad adata_train_gmt.h5ad
The processed data would be saved in adata_train_gmt.h5ad.
python ../script/train.py adata_train_gmt.h5ad model_train Tissue_Source
The trained model would be saved in the model_train/ directory. Tissue_Source is the value of batch_key.
python ../script/predict.py adata_train_gmt.h5ad model_train adata_predict.h5ad adata_predict_gmt.h5ad model_predict
The predicted model would be saved in the model_predict/ directory.
The relevant code could be found in example/example.ipynb and the Application of framework/ directory, including plotting the latent space of the dataset and the influence term score for each cell.