diff --git a/.readthedocs.yml b/.readthedocs.yml
index 9b63ce6dce..8a771612e5 100644
--- a/.readthedocs.yml
+++ b/.readthedocs.yml
@@ -8,7 +8,7 @@ version: 2
# Build documentation in the docs/ directory with Sphinx
sphinx:
builder: html
- configuration: documentation/conf.py
+ configuration: doc/conf.py
fail_on_warning: True
formats:
@@ -16,7 +16,7 @@ formats:
python:
install:
- - requirements: documentation/rtd_requirements.txt
+ - requirements: doc/rtd_requirements.txt
build:
os: "ubuntu-22.04"
apt_packages:
diff --git a/README.md b/README.md
index 7b45537dc3..dd03e6f4f1 100644
--- a/README.md
+++ b/README.md
@@ -1,4 +1,4 @@
- +
+ ## Advanced Multilanguage Interface for CVODES and IDAS
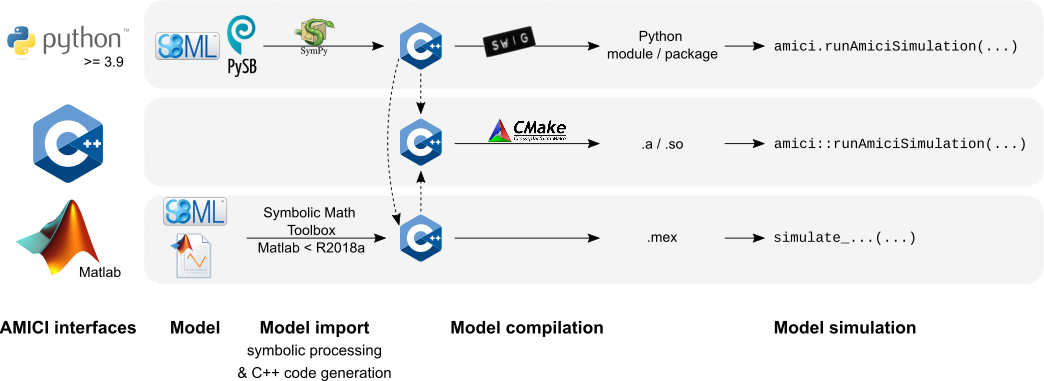
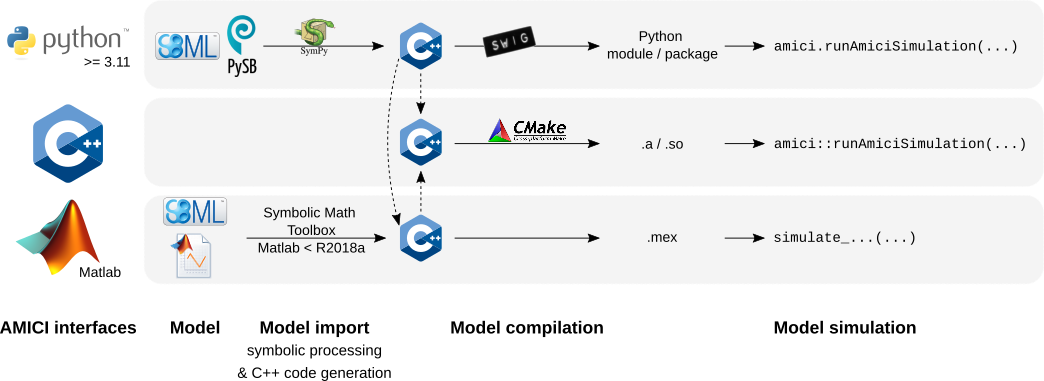
@@ -73,7 +73,7 @@ are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.
-
+
## Getting started
@@ -86,8 +86,8 @@ There are also instructions for using AMICI inside
[containers](https://github.com/AMICI-dev/AMICI/tree/master/container).
To get you started with Python-AMICI, the best way might be checking out this
-[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
-[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
+[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/doc/GettingStarted.ipynb)
+[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=doc%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
@@ -147,11 +147,11 @@ When using AMICI in your project, please cite
```
When presenting work that employs AMICI, feel free to use one of the icons in
-[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
+[doc/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx),
which are available under a
-[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
+[CC0](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx/LICENSE.md)
license:
## Advanced Multilanguage Interface for CVODES and IDAS
@@ -73,7 +73,7 @@ are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.
-
+
## Getting started
@@ -86,8 +86,8 @@ There are also instructions for using AMICI inside
[containers](https://github.com/AMICI-dev/AMICI/tree/master/container).
To get you started with Python-AMICI, the best way might be checking out this
-[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
-[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
+[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/doc/GettingStarted.ipynb)
+[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=doc%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
@@ -147,11 +147,11 @@ When using AMICI in your project, please cite
```
When presenting work that employs AMICI, feel free to use one of the icons in
-[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
+[doc/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx),
which are available under a
-[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
+[CC0](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx/LICENSE.md)
license:
-  +
+ 
diff --git a/binder/overview.ipynb b/binder/overview.ipynb
index 0a7ce81084..b6f915becd 100644
--- a/binder/overview.ipynb
+++ b/binder/overview.ipynb
@@ -8,7 +8,7 @@
"source": [
"# AMICI example notebooks\n",
"\n",
- "* [Getting started](../documentation/GettingStarted.ipynb)\n",
+ "* [Getting started](../doc/GettingStarted.ipynb)\n",
"\n",
" Brief intro to AMICI for first-time users.\n",
"\n",
diff --git a/documentation/CI.md b/doc/CI.md
similarity index 100%
rename from documentation/CI.md
rename to doc/CI.md
diff --git a/documentation/CODE_OF_CONDUCT.md b/doc/CODE_OF_CONDUCT.md
similarity index 100%
rename from documentation/CODE_OF_CONDUCT.md
rename to doc/CODE_OF_CONDUCT.md
diff --git a/documentation/CONTRIBUTING.md b/doc/CONTRIBUTING.md
similarity index 100%
rename from documentation/CONTRIBUTING.md
rename to doc/CONTRIBUTING.md
diff --git a/documentation/CPP.rst b/doc/CPP.rst
similarity index 100%
rename from documentation/CPP.rst
rename to doc/CPP.rst
diff --git a/documentation/CPP_.md b/doc/CPP_.md
similarity index 100%
rename from documentation/CPP_.md
rename to doc/CPP_.md
diff --git a/documentation/ExampleEquilibrationLogic.ipynb b/doc/ExampleEquilibrationLogic.ipynb
similarity index 100%
rename from documentation/ExampleEquilibrationLogic.ipynb
rename to doc/ExampleEquilibrationLogic.ipynb
diff --git a/documentation/ExampleExperimentalConditions.ipynb b/doc/ExampleExperimentalConditions.ipynb
similarity index 100%
rename from documentation/ExampleExperimentalConditions.ipynb
rename to doc/ExampleExperimentalConditions.ipynb
diff --git a/documentation/ExampleJax.ipynb b/doc/ExampleJax.ipynb
similarity index 100%
rename from documentation/ExampleJax.ipynb
rename to doc/ExampleJax.ipynb
diff --git a/documentation/ExampleJaxPEtab.ipynb b/doc/ExampleJaxPEtab.ipynb
similarity index 100%
rename from documentation/ExampleJaxPEtab.ipynb
rename to doc/ExampleJaxPEtab.ipynb
diff --git a/documentation/ExampleSplines.ipynb b/doc/ExampleSplines.ipynb
similarity index 100%
rename from documentation/ExampleSplines.ipynb
rename to doc/ExampleSplines.ipynb
diff --git a/documentation/ExampleSplinesSwameye2003.ipynb b/doc/ExampleSplinesSwameye2003.ipynb
similarity index 100%
rename from documentation/ExampleSplinesSwameye2003.ipynb
rename to doc/ExampleSplinesSwameye2003.ipynb
diff --git a/documentation/ExampleSteadystate.ipynb b/doc/ExampleSteadystate.ipynb
similarity index 100%
rename from documentation/ExampleSteadystate.ipynb
rename to doc/ExampleSteadystate.ipynb
diff --git a/documentation/GettingStarted.ipynb b/doc/GettingStarted.ipynb
similarity index 100%
rename from documentation/GettingStarted.ipynb
rename to doc/GettingStarted.ipynb
diff --git a/documentation/LICENSE.md b/doc/LICENSE.md
similarity index 100%
rename from documentation/LICENSE.md
rename to doc/LICENSE.md
diff --git a/documentation/MATLAB.rst b/doc/MATLAB.rst
similarity index 100%
rename from documentation/MATLAB.rst
rename to doc/MATLAB.rst
diff --git a/documentation/MATLAB_.md b/doc/MATLAB_.md
similarity index 100%
rename from documentation/MATLAB_.md
rename to doc/MATLAB_.md
diff --git a/documentation/PYTHON.rst b/doc/PYTHON.rst
similarity index 100%
rename from documentation/PYTHON.rst
rename to doc/PYTHON.rst
diff --git a/documentation/README.md b/doc/README.md
similarity index 83%
rename from documentation/README.md
rename to doc/README.md
index eece1de90e..f844bc4ea1 100644
--- a/documentation/README.md
+++ b/doc/README.md
@@ -11,13 +11,13 @@ generated using [Sphinx](https://www.sphinx-doc.org/) and related packages.
The legacy GitHub Pages URL https://amici-dev.github.io/AMICI/ is set up as a
redirect to RTD.
-The main configuration file is `documentation/conf.py` and the documentation
+The sphinx configuration file is `doc/conf.py` and the documentation
is generated using `tox -e doc`. The documentation is written to
-`documentation/_build/`.
+`doc/_build/`.
The documentation comprises:
-* reStructuredText / Markdown files from `documentation/`
+* reStructuredText / Markdown files from `doc/`
* Python API documentation of native Python modules
* Python API documentation of Python generated via SWIG (doxygen-style comments
translated to docstrings by SWIG)
@@ -28,14 +28,14 @@ The documentation comprises:
(Parts of the) AMICI documentation can also be directly created using
[doxygen](http://www.doxygen.nl/) directly. It combines Markdown files from
-the root directory, from `documentation/` and in-source documentation from the
-C++ and Matlab source files.
+`doc/` and in-source documentation from the C++ and Matlab source files.
The documentation is generated by running
scripts/run-doxygen.sh
-The resulting HTML and PDF documentation will be created in `doc/`.
+The resulting HTML and PDF documentation will be created in
+`doc/build_doxygen`.
`scripts/run-doxygen.sh` also checks for any missing in-source documentation.
#### Doxygen configuration
@@ -56,8 +56,8 @@ This is configured in `matlab/mtoc/config`.
Out-of-source documentation files should be written in reStructuredText if
intended for Read the Docs or in Markdown if intended for rendering on GitHub.
-Files to be included in the Sphinx/RTD documentation live in `documentation/`.
-Graphics for documentation are kept in `documentation/gfx/`.
+Files to be included in the Sphinx/RTD documentation live in `doc/`.
+Graphics for documentation are kept in `doc/gfx/`.
### When using Markdown
@@ -83,9 +83,9 @@ Graphics for documentation are kept in `documentation/gfx/`.
## Maintaining the list of publications
We want to maintain a list of publications / projects using AMICI. This is
-located at `documentation/references.md`. This file is created by
-`documentation/recreate_reference_list.py` based on
-the bibtex file `documentation/amici_refs.bib`.
+located at `doc/references.md`. This file is created by
+`doc/recreate_reference_list.py` based on
+the bibtex file `doc/amici_refs.bib`.
After any changes to `documentation/amici_refs.bib`, please run
diff --git a/documentation/_templates/autosummary/class.rst b/doc/_templates/autosummary/class.rst
similarity index 100%
rename from documentation/_templates/autosummary/class.rst
rename to doc/_templates/autosummary/class.rst
diff --git a/documentation/_templates/autosummary/module.rst b/doc/_templates/autosummary/module.rst
similarity index 100%
rename from documentation/_templates/autosummary/module.rst
rename to doc/_templates/autosummary/module.rst
diff --git a/documentation/about.rst b/doc/about.rst
similarity index 100%
rename from documentation/about.rst
rename to doc/about.rst
diff --git a/documentation/amici_refs.bib b/doc/amici_refs.bib
similarity index 100%
rename from documentation/amici_refs.bib
rename to doc/amici_refs.bib
diff --git a/documentation/availability.rst b/doc/availability.rst
similarity index 100%
rename from documentation/availability.rst
rename to doc/availability.rst
diff --git a/documentation/background.rst b/doc/background.rst
similarity index 100%
rename from documentation/background.rst
rename to doc/background.rst
diff --git a/documentation/changelog.md b/doc/changelog.md
similarity index 100%
rename from documentation/changelog.md
rename to doc/changelog.md
diff --git a/documentation/code_review_guide.md b/doc/code_review_guide.md
similarity index 100%
rename from documentation/code_review_guide.md
rename to doc/code_review_guide.md
diff --git a/documentation/conf.py b/doc/conf.py
similarity index 98%
rename from documentation/conf.py
rename to doc/conf.py
index 4445c62069..5818cc2bd2 100644
--- a/documentation/conf.py
+++ b/doc/conf.py
@@ -216,7 +216,7 @@ def install_doxygen():
nbsphinx_prolog = (
f"{{% set {ref=} %}}"
r"""
- {% set docname = "documentation/" + env.doc2path(env.docname, base=False)|string %}
+ {% set docname = "doc/" + env.doc2path(env.docname, base=False)|string %}
.. raw:: html
@@ -600,9 +600,9 @@ def process_signature(
# this code fixes references in symlinked md files in documentation folder
# link replacements must be in env.domains['std'].labels
doclinks = {
- "documentation/development": "/development.md",
- "documentation/CI": "/ci.md",
- "documentation/code_review_guide": "/code_review_guide.md",
+ "doc/development": "/development.md",
+ "doc/CI": "/ci.md",
+ "doc/code_review_guide": "/code_review_guide.md",
}
diff --git a/documentation/contributing.rst b/doc/contributing.rst
similarity index 100%
rename from documentation/contributing.rst
rename to doc/contributing.rst
diff --git a/documentation/cpp_installation.rst b/doc/cpp_installation.rst
similarity index 100%
rename from documentation/cpp_installation.rst
rename to doc/cpp_installation.rst
diff --git a/documentation/cpp_interface.rst b/doc/cpp_interface.rst
similarity index 100%
rename from documentation/cpp_interface.rst
rename to doc/cpp_interface.rst
diff --git a/documentation/debugging.rst b/doc/debugging.rst
similarity index 100%
rename from documentation/debugging.rst
rename to doc/debugging.rst
diff --git a/documentation/development.rst b/doc/development.rst
similarity index 100%
rename from documentation/development.rst
rename to doc/development.rst
diff --git a/documentation/example_errors.ipynb b/doc/example_errors.ipynb
similarity index 100%
rename from documentation/example_errors.ipynb
rename to doc/example_errors.ipynb
diff --git a/documentation/example_large_models b/doc/example_large_models
similarity index 100%
rename from documentation/example_large_models
rename to doc/example_large_models
diff --git a/documentation/gfx/LICENSE.md b/doc/gfx/LICENSE.md
similarity index 100%
rename from documentation/gfx/LICENSE.md
rename to doc/gfx/LICENSE.md
diff --git a/documentation/gfx/README.md b/doc/gfx/README.md
similarity index 100%
rename from documentation/gfx/README.md
rename to doc/gfx/README.md
diff --git a/documentation/gfx/amici_workflow.png b/doc/gfx/amici_workflow.png
similarity index 100%
rename from documentation/gfx/amici_workflow.png
rename to doc/gfx/amici_workflow.png
diff --git a/documentation/gfx/amici_workflow.svg b/doc/gfx/amici_workflow.svg
similarity index 100%
rename from documentation/gfx/amici_workflow.svg
rename to doc/gfx/amici_workflow.svg
diff --git a/documentation/gfx/banner.png b/doc/gfx/banner.png
similarity index 100%
rename from documentation/gfx/banner.png
rename to doc/gfx/banner.png
diff --git a/documentation/gfx/logo.png b/doc/gfx/logo.png
similarity index 100%
rename from documentation/gfx/logo.png
rename to doc/gfx/logo.png
diff --git a/documentation/gfx/logo_template.svg b/doc/gfx/logo_template.svg
similarity index 100%
rename from documentation/gfx/logo_template.svg
rename to doc/gfx/logo_template.svg
diff --git a/documentation/gfx/logo_text.png b/doc/gfx/logo_text.png
similarity index 100%
rename from documentation/gfx/logo_text.png
rename to doc/gfx/logo_text.png
diff --git a/documentation/gfx/steadystate_solver_workflow.png b/doc/gfx/steadystate_solver_workflow.png
similarity index 100%
rename from documentation/gfx/steadystate_solver_workflow.png
rename to doc/gfx/steadystate_solver_workflow.png
diff --git a/documentation/glossary.rst b/doc/glossary.rst
similarity index 100%
rename from documentation/glossary.rst
rename to doc/glossary.rst
diff --git a/documentation/how_to_cite.rst b/doc/how_to_cite.rst
similarity index 100%
rename from documentation/how_to_cite.rst
rename to doc/how_to_cite.rst
diff --git a/documentation/implementation_discontinuities.rst b/doc/implementation_discontinuities.rst
similarity index 100%
rename from documentation/implementation_discontinuities.rst
rename to doc/implementation_discontinuities.rst
diff --git a/documentation/index.rst b/doc/index.rst
similarity index 100%
rename from documentation/index.rst
rename to doc/index.rst
diff --git a/documentation/matlab_faq.rst b/doc/matlab_faq.rst
similarity index 100%
rename from documentation/matlab_faq.rst
rename to doc/matlab_faq.rst
diff --git a/documentation/matlab_installation.rst b/doc/matlab_installation.rst
similarity index 100%
rename from documentation/matlab_installation.rst
rename to doc/matlab_installation.rst
diff --git a/documentation/matlab_interface.rst b/doc/matlab_interface.rst
similarity index 100%
rename from documentation/matlab_interface.rst
rename to doc/matlab_interface.rst
diff --git a/documentation/model_steadystate_scaled.xml b/doc/model_steadystate_scaled.xml
similarity index 100%
rename from documentation/model_steadystate_scaled.xml
rename to doc/model_steadystate_scaled.xml
diff --git a/documentation/model_steadystate_scaled_without_observables.xml b/doc/model_steadystate_scaled_without_observables.xml
similarity index 100%
rename from documentation/model_steadystate_scaled_without_observables.xml
rename to doc/model_steadystate_scaled_without_observables.xml
diff --git a/documentation/petab.ipynb b/doc/petab.ipynb
similarity index 100%
rename from documentation/petab.ipynb
rename to doc/petab.ipynb
diff --git a/documentation/python_examples.rst b/doc/python_examples.rst
similarity index 100%
rename from documentation/python_examples.rst
rename to doc/python_examples.rst
diff --git a/documentation/python_faq.rst b/doc/python_faq.rst
similarity index 100%
rename from documentation/python_faq.rst
rename to doc/python_faq.rst
diff --git a/documentation/python_installation.rst b/doc/python_installation.rst
similarity index 100%
rename from documentation/python_installation.rst
rename to doc/python_installation.rst
diff --git a/documentation/python_interface.rst b/doc/python_interface.rst
similarity index 100%
rename from documentation/python_interface.rst
rename to doc/python_interface.rst
diff --git a/documentation/python_modules.rst b/doc/python_modules.rst
similarity index 100%
rename from documentation/python_modules.rst
rename to doc/python_modules.rst
diff --git a/documentation/recreate_reference_list.py b/doc/recreate_reference_list.py
similarity index 100%
rename from documentation/recreate_reference_list.py
rename to doc/recreate_reference_list.py
diff --git a/documentation/references.md b/doc/references.md
similarity index 100%
rename from documentation/references.md
rename to doc/references.md
diff --git a/documentation/rtd_requirements.txt b/doc/rtd_requirements.txt
similarity index 100%
rename from documentation/rtd_requirements.txt
rename to doc/rtd_requirements.txt
diff --git a/documentation/versioning_policy.rst b/doc/versioning_policy.rst
similarity index 100%
rename from documentation/versioning_policy.rst
rename to doc/versioning_policy.rst
diff --git a/matlab/mtoc/config/Doxyfile.template b/matlab/mtoc/config/Doxyfile.template
index f5eed70f73..b59cff3cdf 100644
--- a/matlab/mtoc/config/Doxyfile.template
+++ b/matlab/mtoc/config/Doxyfile.template
@@ -947,8 +947,8 @@ WARN_LOGFILE =
INPUT = "_SourceDir_/README.md" \
"_SourceDir_/LICENSE.md" \
- "_SourceDir_/documentation/MATLAB_.md" \
- "_SourceDir_/documentation/CPP_.md" \
+ "_SourceDir_/doc/MATLAB_.md" \
+ "_SourceDir_/doc/CPP_.md" \
"_SourceDir_/include" \
"_SourceDir_/matlab"
@@ -1403,15 +1403,6 @@ HTML_COLORSTYLE_SAT = 100
HTML_COLORSTYLE_GAMMA = 80
-# If the HTML_TIMESTAMP tag is set to YES then the footer of each generated HTML
-# page will contain the date and time when the page was generated. Setting this
-# to YES can help to show when doxygen was last run and thus if the
-# documentation is up to date.
-# The default value is: NO.
-# This tag requires that the tag GENERATE_HTML is set to YES.
-
-HTML_TIMESTAMP = YES
-
# If the HTML_DYNAMIC_MENUS tag is set to YES then the generated HTML
# documentation will contain a main index with vertical navigation menus that
# are dynamically created via JavaScript. If disabled, the navigation index will
@@ -2072,14 +2063,6 @@ LATEX_HIDE_INDICES = YES
LATEX_BIB_STYLE = plain
-# If the LATEX_TIMESTAMP tag is set to YES then the footer of each generated
-# page will contain the date and time when the page was generated. Setting this
-# to NO can help when comparing the output of multiple runs.
-# The default value is: NO.
-# This tag requires that the tag GENERATE_LATEX is set to YES.
-
-LATEX_TIMESTAMP = NO
-
# The LATEX_EMOJI_DIRECTORY tag is used to specify the (relative or absolute)
# path from which the emoji images will be read. If a relative path is entered,
# it will be relative to the LATEX_OUTPUT directory. If left blank the
diff --git a/python/examples/example_constant_species/ExampleEquilibrationLogic.ipynb b/python/examples/example_constant_species/ExampleEquilibrationLogic.ipynb
index dc0bf6afed..d8c5a2f90e 100644
--- a/python/examples/example_constant_species/ExampleEquilibrationLogic.ipynb
+++ b/python/examples/example_constant_species/ExampleEquilibrationLogic.ipynb
@@ -65,9 +65,7 @@
"source": [
"from IPython.display import Image\n",
"\n",
- "fig = Image(\n",
- " filename=\"../../../documentation/gfx/steadystate_solver_workflow.png\"\n",
- ")\n",
+ "fig = Image(filename=\"../../../doc/gfx/steadystate_solver_workflow.png\")\n",
"fig"
]
},
diff --git a/python/examples/example_large_models/example_performance_optimization.ipynb b/python/examples/example_large_models/example_performance_optimization.ipynb
index 82ca8d9dbb..89c7552d83 100644
--- a/python/examples/example_large_models/example_performance_optimization.ipynb
+++ b/python/examples/example_large_models/example_performance_optimization.ipynb
@@ -19,7 +19,7 @@
"4. Compiling the generated code\n",
"5. Simulating the model\n",
"\n",
- "\n",
+ "\n",
"\n",
"There are various options to speed up individual steps of this process. Generally, faster import comes with slower simulation and vice versa. During parameter estimation, a model is often imported only once, and then millions of simulations are run. Therefore, faster simulation will easily compensate for slower import (one-off cost). In other cases, many models may to have to be imported, but only few simulations will be executed. In this case, faster import may be more relevant.\n",
"\n",
@@ -308,7 +308,7 @@
"#### Parallel compilation\n",
"\n",
"It's possible to compile multiple source files in parallel by specifying the number of parallel processes via the `AMICI_PARALLEL_COMPILE` environment variable. This is also beneficial for small models.\n",
- "Note, however, that for large models, this may require significant amounts of RAM. \n",
+ "Note, however, that for large models, this may require significant amounts of RAM.\n",
"\n",
"Example for a large and tiny model:"
]
@@ -388,7 +388,7 @@
"\n",
"Another potential performance gain can be obtained from using CPU-specific instructions using `-march=native`. The disadvantage is, that the compiled model extension will only run on CPUs supporting the same instruction set. This may be become problematic when attempting to use an AMICI model on a machine other than on which it was compiled (e.g. on heterogeneous compute clusters).\n",
"\n",
- "These compiler flags should be set for both, AMICI installation and model compilation. \n",
+ "These compiler flags should be set for both, AMICI installation and model compilation.\n",
"\n",
"For AMICI installation, e.g.,\n",
"```bash\n",
@@ -485,7 +485,7 @@
"source": [
"## Model simulation\n",
"\n",
- "A major determinant of simulation time for a given model is the required accuracy and the selected solvers. This has been evaluated, for example, in https://doi.org/10.1038/s41598-021-82196-2 and is not covered further here. \n",
+ "A major determinant of simulation time for a given model is the required accuracy and the selected solvers. This has been evaluated, for example, in https://doi.org/10.1038/s41598-021-82196-2 and is not covered further here.\n",
"\n",
"### Adjoint *vs.* forward sensitivities\n",
"\n",
@@ -513,7 +513,7 @@
"\n",
"### Reporting mode\n",
"\n",
- "During model simulation, many quantities are calculated, but not all might be of interest for you. For example, for parameter estimation you might only be interested in the likelihood and gradient. In this case, you can save time and memory using \n",
+ "During model simulation, many quantities are calculated, but not all might be of interest for you. For example, for parameter estimation you might only be interested in the likelihood and gradient. In this case, you can save time and memory using\n",
"[amici.Solver.setReturnDataReportingMode(amici.RDataReporting.likelihood)](https://amici.readthedocs.io/en/latest/generated/amici.amici.Solver.html#amici.amici.Solver.setReturnDataReportingMode)."
]
}
diff --git a/scripts/run-doxygen.sh b/scripts/run-doxygen.sh
index c766d93eed..0b9f78b33f 100755
--- a/scripts/run-doxygen.sh
+++ b/scripts/run-doxygen.sh
@@ -6,6 +6,9 @@ set -x
SCRIPT_PATH=$(dirname $BASH_SOURCE)
AMICI_PATH=$(cd $SCRIPT_PATH/.. && pwd)
+OUTDIR="${AMICI_PATH}/doc/build_doxygen"
+mkdir -p "$OUTDIR"
+
# Set up Matlab filter
cd ${AMICI_PATH}
scripts/downloadAndBuildMtocpp.sh
@@ -17,7 +20,7 @@ cp "${MTOC_CONFIG_PATH}/Doxyfile.template" "${DOXYFILE}"
DOXY_WARNING_FILE=${AMICI_PATH}/matlab/mtoc/warnings.log
# Replace some template values
-sed -i -e "s#_OutputDir_#$AMICI_PATH/doc#g" ${DOXYFILE}
+sed -i -e "s#_OutputDir_#$OUTDIR#g" ${DOXYFILE}
sed -i -e "s#_SourceDir_#$AMICI_PATH#g" ${DOXYFILE}
sed -i -e "s#_ConfDir_#${MTOC_CONFIG_PATH}#g" ${DOXYFILE}
sed -i -e "s#_ProjectName_#AMICI#g" ${DOXYFILE}
@@ -45,7 +48,7 @@ rm "${DOXYFILE}"
rm "${MTOC_CONFIG_PATH}/mtocpp_filter.sh"
# Build pdf
-cd "${AMICI_PATH}/doc/latex"
+cd "${OUTDIR}/latex"
make
cp ./refman.pdf "${AMICI_PATH}/AMICI_guide.pdf"
diff --git a/tox.ini b/tox.ini
index 9f060a7cf0..4c10b020dd 100644
--- a/tox.ini
+++ b/tox.ini
@@ -12,10 +12,10 @@ passenv = AMICI_PARALLEL_COMPILE,CC,CXX
description =
Build documentation
deps =
- -r documentation/rtd_requirements.txt
+ -r doc/rtd_requirements.txt
# don't install the package, this is already handled by `deps` above
skip_install = true
-change_dir = documentation/
+change_dir = doc/
allowlist_externals =
rm
commands =
 +
+ 
 +
+ ## Advanced Multilanguage Interface for CVODES and IDAS
@@ -73,7 +73,7 @@ are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.
-
+
## Getting started
@@ -86,8 +86,8 @@ There are also instructions for using AMICI inside
[containers](https://github.com/AMICI-dev/AMICI/tree/master/container).
To get you started with Python-AMICI, the best way might be checking out this
-[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
-[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
+[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/doc/GettingStarted.ipynb)
+[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=doc%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
@@ -147,11 +147,11 @@ When using AMICI in your project, please cite
```
When presenting work that employs AMICI, feel free to use one of the icons in
-[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
+[doc/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx),
which are available under a
-[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
+[CC0](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx/LICENSE.md)
license:
## Advanced Multilanguage Interface for CVODES and IDAS
@@ -73,7 +73,7 @@ are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.
-
+
## Getting started
@@ -86,8 +86,8 @@ There are also instructions for using AMICI inside
[containers](https://github.com/AMICI-dev/AMICI/tree/master/container).
To get you started with Python-AMICI, the best way might be checking out this
-[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
-[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
+[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/doc/GettingStarted.ipynb)
+[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=doc%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
@@ -147,11 +147,11 @@ When using AMICI in your project, please cite
```
When presenting work that employs AMICI, feel free to use one of the icons in
-[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
+[doc/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx),
which are available under a
-[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
+[CC0](https://github.com/AMICI-dev/AMICI/tree/master/doc/gfx/LICENSE.md)
license:
 +
+ 